Difference between revisions of "Weighted Least Squares (WLS)"
(→Definition of local aproximation) |
(→Definition of local aproximation) |
||
| Line 5: | Line 5: | ||
</figure> | </figure> | ||
== Definition of local aproximation == | == Definition of local aproximation == | ||
| − | Our wish is to approximate an unknown function $u\colon \R^k \to \R$ while knowing $n$ values $u(\vec{ | + | Our wish is to approximate an unknown function $u\colon \R^k \to \R$ while knowing $n$ values $u(\vec{s}_j) := u_j$. |
The vector of known values will be denoted by $\b{u}$ and the vector of coordinates where those values were achieved by $\b{s}$. | The vector of known values will be denoted by $\b{u}$ and the vector of coordinates where those values were achieved by $\b{s}$. | ||
Note that $\b{x}$ is not a vector in the usual sense since its components $\vec{x}_j$ are elements of $\R^k$, but we will call it vector anyway. | Note that $\b{x}$ is not a vector in the usual sense since its components $\vec{x}_j$ are elements of $\R^k$, but we will call it vector anyway. | ||
Revision as of 12:09, 24 October 2016
One of the most important building blocks of the meshless methods is the Moving Least Squares approximation, which is implemented in the EngineMLS class. Check EngineMLS unit tests for examples.
Contents
Definition of local aproximation
Our wish is to approximate an unknown function $u\colon \R^k \to \R$ while knowing $n$ values $u(\vec{s}_j) := u_j$. The vector of known values will be denoted by $\b{u}$ and the vector of coordinates where those values were achieved by $\b{s}$. Note that $\b{x}$ is not a vector in the usual sense since its components $\vec{x}_j$ are elements of $\R^k$, but we will call it vector anyway. The values of $\b{x}$ are called nodes or support nodes or support. The known values $\bf{u}$ are also called support values.
In general, an approximation function around point $\vec{p}\in\R^k$ can be written as \[\hat{u} (\vec{x}) = \sum_{i=1}^m \alpha_i b_i(\vec{x}) = \b{b}(\vec{x})^\T \b{\alpha} \] where $\b{b} = (b_i)_{i=1}^m$ is a set of basis functions, $b_i\colon \R^k \to\R$, and $\b{\alpha} = (\alpha_i)_{i=1}^m$ are the unknown coefficients.
In MLS the goal is to minimize the error of approximation in given values, $\b{e} = \hat u(\b{s}) - \b{u}$ between the approximation function and target function in the known points $\b{x}$. The error can also be written as $B\b{\alpha} - \b{u}$, where $B$ is rectangular matrix of dimensions $n \times m$ with rows containing basis function evaluated in points $\vec{s}_j$. \[ B = \begin{bmatrix} b_1(\vec{s_1}) & \ldots & b_m(\vec{s_1}) \\ \vdots & \ddots & \vdots \\ b_1(\vec{s_n}) & \ldots & b_m(\vec{s_n}) \end{bmatrix} = [b_i(\vec{s}_j)]_{i=1,j=1}^{m,n} = [\b{b}(\vec{s}_j)^\T]_{j=1}^n. \]
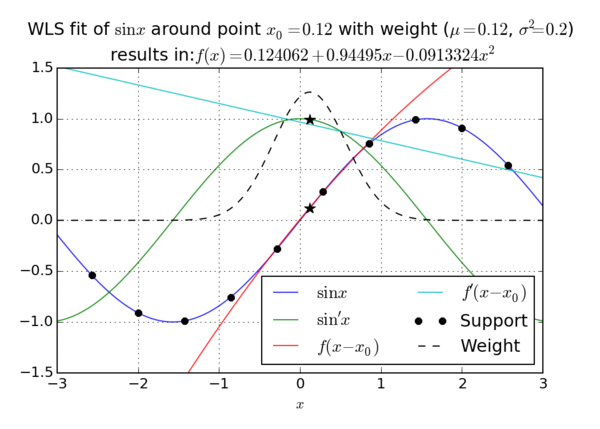
We can choose to minimize any norm of the error vector $e$ and usually choose to minimize the 2-norm or square norm \[ \|\b{e}\| = \|\b{e}\|_2 = \sqrt{\sum_{j=1}^n e_j^2}. \] Commonly, we also choose to minimize a weighted norm [1] instead \[ \|\b{e}\|_{2,w} = \|\b{e}\|_w = \sqrt{\sum_{j=1}^n (w_j e_j)^2}. \] The weights $w_i$ are assumed to be non negative and are assembled in a vector $\b{w}$ or a matrix $W = \operatorname{diag}(\b{w})$ and usually obtained from a weight function. A weight function is a function $\omega\colon \R^k \to[0,\infty)$. We calculate $w_j$ as $w_i := \omega(\vec{p}-\vec{s}_j)$, so good choices for $\omega$ are function which have higher values close to 0 (making closer nodes more important), like the normal distribution. If we choose $\omega \equiv 1$, we get the unweighted version.
A choice of minimizing the square norm gave this method its name - Least Squares appoximation. If we use the weighted version, we get the Weighted Least Squares or WLS. In the most general case we wish to minimize \[ \|\b{e}\|_{2,w}^2 = \b{e}^\T W^2 \b{e} = (B\b{\alpha} - \b{u})^\T W^2(B\b{\alpha} - \b{u}) = \sum_j^n w_j^2 (\hat{u}(\vec{s}_j) - u_j)^2 \]
The problem of finding the coefficients $\b{\alpha}$ that minimize the error $\b{e}$ can be solved with at least three approaches:
- Normal equations (fastest, less accurate) - using Cholesky decomposition of $B$ (requires full rank and $m \leq n$)
- QR decomposition of $B$ (requires full rank and $m \leq n$, more precise)
- SVD decomposition of $B$ (more expensive, even more reliable, no rank demand)
In MM we use SVD with regularization described below.
Computing approximation coefficients
Normal equations
We seek the minimum of \[ \|\b{e}\|_2^2 = (B\b{\alpha} - \b{u})^\T(B\b{\alpha} - \b{u}) \] By seeking the zero gradient in terms of coefficients $\alpha_i$ \[\frac{\partial}{\partial \alpha_i} (B\b{\alpha} - \b{u})^\T(B\b{\alpha} - \b{u}) = 0\] resulting in \[ B^\T B\b{\alpha} = B^\T \b{u}. \] The coefficient matrix $B^\T B$ is symmetric and positive definite. However, solving above problem directly is poorly behaved with respect to round-off errors since the condition number $\kappa(B^\T B)$ is the square of $\kappa(B)$.
In case of WLS the equations become \[ (WB)^\T WB \b{\alpha} = WB^\T \b{u}. \]
Complexity of Cholesky decomposition is $\frac{n^3}{3}$ and complexity of matrix multiplication $nm^2$. To preform the Cholesky decomposition the rank of $WB$ must be full.
Pros:
- simple to implement
- low computational complexity
Cons:
- numerically unstable
- full rank requirement
QR Decomposition
\[{\bf{B}} = {\bf{QR}} = \left[ {{{\bf{Q}}_1},{{\bf{Q}}_2}} \right]\left[ {\begin{array}{*{20}{c}} {{{\bf{R}}_1}}\\ 0 \end{array}} \right]\] \[{\bf{B}} = {{\bf{Q}}_1}{{\bf{R}}_1}\] $\bf{Q}$ is unitary matrix ($\bf{Q}^{-1}=\bf{Q}^T$). Useful property of a unitary matrices is that multiplying with them does not alter the (Euclidean) norm of a vector, i.e., \[\left\| {{\bf{Qx}}} \right\|{\bf{ = }}\left\| {\bf{x}} \right\|\] And $\bf{R}$ is upper diagonal matrix \[{\bf{R = (}}{{\bf{R}}_{\bf{1}}}{\bf{,}}0{\bf{)}}\] therefore we can say \[\begin{array}{l} \left\| {{\bf{B\alpha }} - {\bf{u}}} \right\| = \left\| {{{\bf{Q}}^{\rm{T}}}\left( {{\bf{B\alpha }} - {\bf{u}}} \right)} \right\| = \left\| {{{\bf{Q}}^{\rm{T}}}{\bf{B\alpha }} - {{\bf{Q}}^{\rm{T}}}{\bf{u}}} \right\|\\ = \left\| {{{\bf{Q}}^{\rm{T}}}\left( {{\bf{QR}}} \right){\bf{\alpha }} - {{\bf{Q}}^{\rm{T}}}{\bf{u}}} \right\| = \left\| {\left( {{{\bf{R}}_1},0} \right){\bf{\alpha }} - {{\left( {{{\bf{Q}}_1},{{\bf{Q}}_{\bf{2}}}} \right)}^{\rm{T}}}{\bf{u}}} \right\|\\ = \left\| {{{\bf{R}}_{\bf{1}}}{\bf{\alpha }} - {{\bf{Q}}_{\bf{1}}}{\bf{u}}} \right\| + \left\| {{\bf{Q}}_2^{\rm{T}}{\bf{u}}} \right\| \end{array}\] Of the two terms on the right we have no control over the second, and we can render the first one zero by solving \[{{\bf{R}}_{\bf{1}}}{\bf{\alpha }} = {\bf{Q}}_{_{\bf{1}}}^{\rm{T}}{\bf{u}}\] Which results in a minimum. We could also compute it with pseudo inverse \[\mathbf{\alpha }={{\mathbf{B}}^{-1}}\mathbf{u}\] Where pseudo inverse is simply \[{{\mathbf{B}}^{-1}}=\mathbf{R}_{\text{1}}^{\text{-1}}{{\mathbf{Q}}^{\text{T}}}\] (once again, R is upper diagonal matrix, and Q is unitary matrix). And for weighted case \[\mathbf{\alpha }={{\left( {{\mathbf{W}}^{0.5}}\mathbf{B} \right)}^{-1}}\left( {{\mathbf{W}}^{0.5}}\mathbf{u} \right)\]
Complexity of QR decomposition \[\frac{2}{3}m{{n}^{2}}+{{n}^{2}}+\frac{1}{3}n-2=O({{n}^{3}})\]
Pros: better stability in comparison with normal equations cons: higher complexity
SVD decomposition
In linear algebra, the singular value decomposition (SVD) is a factorization of a real or complex matrix. It has many useful applications in signal processing and statistics.
Formally, the singular value decomposition of an $m \times n$ real or complex matrix $\bf{B}$ is a factorization of the form $\bf{B}= \bf{U\Sigma V^\T}$, where $\bf{U}$ is an $m \times m$ real or complex unitary matrix, $\bf{\Sigma}$ is an $m \times n$ rectangular diagonal matrix with non-negative real numbers on the diagonal, and $\bf{V}^\T$ is an $n \times n$ real or complex unitary matrix. The diagonal entries $\Sigma_{ii}$ are known as the singular values of $\bf{B}$. The $m$ columns of $\bf{U}$ and the $n$ columns of $\bf{V}$ are called the left-singular vectors and right-singular vectors of $\bf{B}$, respectively.
The singular value decomposition and the eigen decomposition are closely related. Namely:
- The left-singular vectors of $\bf{B}$ are eigen vectors of $\bf{BB}^\T$.
- The right-singular vectors of $\bf{B}$ are eigen vectors of $\bf{B}^\T{B}$.
- The non-zero singular values of $\bf{B}$ (found on the diagonal entries of $\bf{\Sigma}$) are the square roots of the non-zero eigenvalues of both $\bf{B}^\T\bf{B}$ and $\bf{B}^\T \bf{B}$.
with SVD we can write \bf{B} as \[\bf{B}=\bf{U\Sigma{{V}^{\T}}}\] where are \bf{U} and \bf{V} again unitary matrices and $\bf{\Sigma}$ stands for diagonal matrix of singular values.
Again we can solve either the system or compute pseudo inverse as
\[ \bf{B}^{-1} = \left( \bf{U\Sigma V}^\T\right)^{-1} = \bf{V}\bf{\Sigma^{-1}U}^\T \] where $\bf{\Sigma}^{-1}$ is trivial, just replace every non-zero diagonal entry by its reciprocal and transpose the resulting matrix. The stability gain is exactly here, one can now set threshold below which the singular value is considered as 0, basically truncate all singular values below some value and thus stabilize the inverse.
SVD decomposition complexity \[ 2mn^2+2n^3 = O(n^3) \]
Pros: stable cons: high complexity
Method used in MM (SVD with regularization)
Weighted Least Squares
TODO
WLS at fixed point with fixed support and unknown function values
Blah blah, shape funkcije ... TODO
Moving Least Squares
TODO
End notes
- ↑ Note that our definition is a bit unusual, usually weights are not squared with the values. However, we do this to avoid computing square roots when doing MLS. If you are used to the usual definition, consider the weight to be $\omega^2$.