Positioning of computational nodes
Although the meshless methods do not require any topological relations between nodes and even randomly distributed nodes could be used, it is well-known that using regularly distributed nodes leads to more accurate and more stable results. So despite meshless seeming robustness regarding the nodal distribution, a certain effort has to be invested into the positioning of the nodes and following discussion, to some extent, deals with this problem. In a following discussion we want to form set of algorithms that can cover arbitrary domain in n dimensions with nodes that can be further used to solve systems of PDEs. We start with algorithms for filling the domain with nodes. Second set of algorithms is denoted to improving of the nodal distributions and a last set of algorithms deals with the refinement of the nodal distribution. The algorithms can of course be combined.
Contents
Filling domain with nodes
The goal is to fill arbitrary domain, which we can ask if position $\b{p}$ is in the domain our outside, with nodes following the given target density function $\rho(\b{p})$.
We start with a simple algorithm based on Poisson Disk Sampling (PDS) that results in a relatively tightly packed distribution of nodes. What we do is:
- Randomly select a node within the domain.
- Add N (depending on the dimensionality of the space) nodes on circle/sphere around that node.
- Randomly select one of just added nodes and repeat adding.
- Each time check if any of new nodes violates minimal distance criterion, i.e. if it is positioned to close to any of the existing nodes.
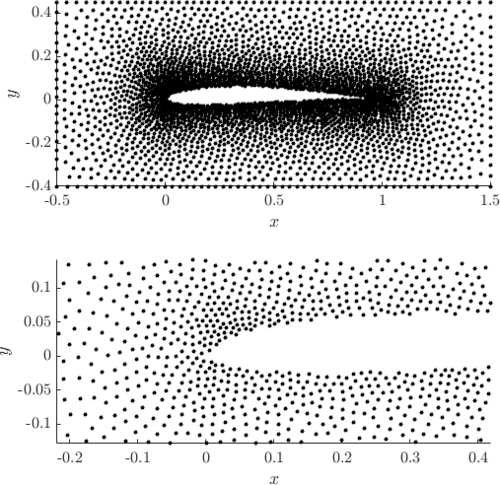
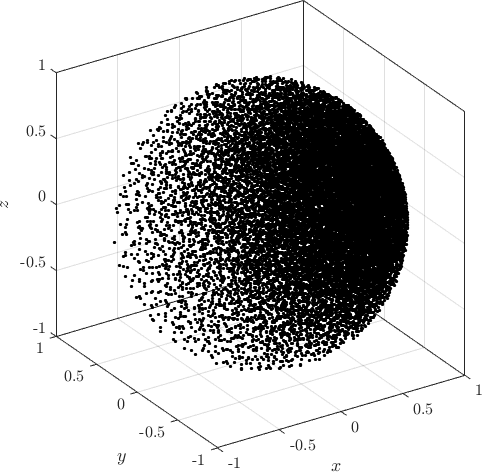
Examples of PDS discretized irregular domains in 2D (first code snippet, left image) and in 3D (second code snippet, right image) can be found below.
1 // setup
2 PoissonDiskSamplingFill fill_engine;
3 fill_engine.iterations(1000000).optim_after(1000).use_MC(true).proximity_relax(0.8);
4 // create test domain
5 RectangleDomain<Vec2d> domain({0, 0}, {1.2, 1.5});
6 CircleDomain<Vec2d> o1({0.4, 0.4}, 0.25);
7 domain.subtract(o1);
8 // define density function
9 auto fill_density = [](Vec2d p)->double{
10 return (std::pow(p[0]/10, 2) + 0.01);
11 };
12 fill_engine(domain, fill_density);
1 PoissonDiskSamplingFill fill_engine;
2 // setup
3 PoissonDiskSamplingFill fill_engine;
4 fill_engine.iterations(1000000).optim_after(1000).use_MC(true).proximity_relax(0.8);
5 // create test domain
6 RectangleDomain<Vec3d> domain({0, 0, 0}, {1.2, 1.5, 1.1});
7 CircleDomain<Vec3d> o1({0, 0, 0}, 0.5);
8 domain.subtract(o1);
9 // define target density
10 auto fill_density = [](Vec3d p)->double{
11 return (std::pow(p[0]/10 + p[1]/10 + p[2]/10, 2) + 0.025);
12 };
13 fill_engine(domain, fill_density);
Examples of PDS discretized in 2D and in 3D generated with above code.
Relaxation of the nodal distribution
To construct stable and reliable shape functions the support domains need to be non-degenerated [1], i.e. the distances between support nodes have to be balanced. Naturally, this condition is fulfilled in regular nodal distributions, but when working with complex geometries, the nodes have to be positioned accordingly. There are different algorithms designed to optimally fill the domain with different shapes [2, 3]. Here an intrinsic feature of the MLSM is used to take care of that problem. The goal is to minimize the overall support domain degeneration in order to attain stable numerical solution. In other words, a global optimization problem with the overall deformation of the local support domains acting as the cost function is tackled. We seek the global minimum by a local iterative approach. In each iteration, the computational nodes are translated according to the local derivative of the potential \begin{equation} \delta \b{p}\left( \b{p} \right)=-\sigma_{k}\sum\limits_{n=1}^{n}{\nabla }V\left( \mathbf{p}-\b{p}_n \right) \end{equation} where $V, n, \delta \b{p}, \b{p}_{n}$ and $\sigma_{k}$ stand for the potential, number of support nodes, offset of the node, position of n-th support node and relaxation parameter, respectively. After offsets in all nodes are computed, the nodes are repositioned as $\b{p}\leftarrow \b{p}+\delta \b{p}\left( \b{p} \right)$. Presented iterative process procedure begins with positioning of boundary nodes, which is considered as the definition of the domain, and then followed by the positioning of internal nodes.
The BasicRelax Engine supports two call types:
- with supplied distribution function relax(func), where it tries to satisfy the user supplied nodal density function. This can be achieved only when there is the total number of domain nodes the same as integral of density function over the domain. If there is too much nodes a volatile relax might occur. If there is not enough nodes the relax might become lazy. The best use of this mode is in combination with fillDistribution Engines.
- without distribution, where nodes always move towards less populated area. The relax magnitude is simply determined from Annealing factor and distance to the closest node. A simple and stable approach, however, note that this relax always converges towards uniformly distributed nodes.
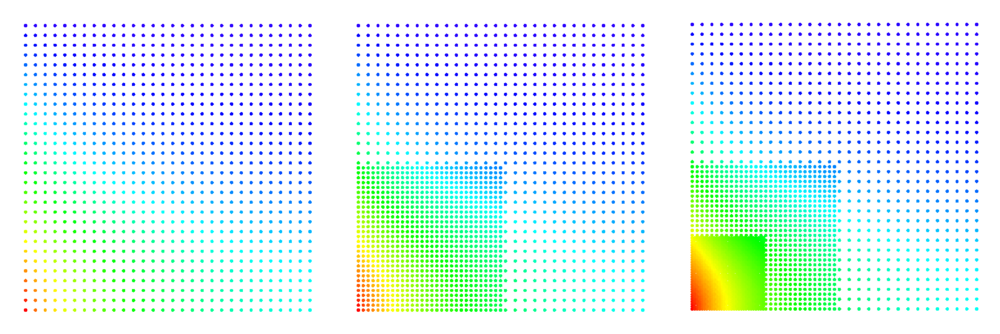
Example of filling and relaxing 2D domain can be found in below code snippet and Figure. Note the difference between relax without supplied distribution (right) and with supplied distribution (left). The quiver plot represents normal vectors in boundary nodes. More examples can be found in main repository under tests.
1 double r = 0.25;
2 CircleDomain<Vec2d> c({0.5, 0.5}, r);
3
4 BasicRelax relax;
5 relax.iterations(100).InitialHeat(1).FinalHeat(0).projectionType(1).numNeighbours(3);
6 relax.boundaryProjectionThreshold(0.55);
7 auto fill_density = [](Vec2d p) -> double {
8 return (0.005 + (p[0] - 0.5) * (p[0] - 0.5) / 2 + (p[1] - 0.5) * (p[1] - 0.5) / 2);
9 };
10
11 c.fillUniformBoundaryWithStep(fill_density(Vec2d({r, 0.0})));
12 PoissonDiskSamplingFill fill_engine;
13
14 fill_engine(c, fill_density);
15 relax(c);
[1] Lee CK, Liu X, Fan SC. Local muliquadric approximation for solving boundary value problems. Comput Mech. 2003;30:395-409.
[2] Löhner R, Oñate E. A general advancing front technique for filling space with arbitrary objects. Int J Numer Meth Eng. 2004;61:1977-91.
[3] Liu Y, Nie Y, Zhang W, Wang L. Node placement method by bubble simulation and its application. CMES-Comp Model Eng. 2010;55:89.
Refinement of the nodal distribution
Here we consider possible meshless refinement algorithms (sometimes also called adaptive cloud refinement). The refinement mechanisms we have so far studied include:
- refinement based on closest node distance
- refinement based on averaged (inter-)node distance
- refinement based on half-links
Here we only want to compare the quality of the refined grids and have not tied the refinement algorithm with a error indicator, thus we only study the node insertion process by refining the whole grid.
The refinement routine takes a range of nodes (e.g. a subregion of the domain) together with the refinement parameters and generates new nodes around the old ones. Special care must be taken with refinement of the boundary nodes. Points have to be selected on the actual boundary either analytically considering the geometry or with a numerical root finder such as bisection.
Problem description
To compare the node refinement mechanisms we study the process of reaction-diffusion in an infinite cylindrical catalyst pellet (infinite in the $z$-dimension). Since the pellet is infinite in one dimension this problem simplifies to a 2D problem (in the $xy$-plane). For a catalyst pellet of radius $R$ centered at $(x,y) = (0,0)$ and the reactant undergoing a first order reaction we must solve the equation \begin{equation} \b{\nabla}^2 C - {M_T}^2 C = 0, \end{equation} where $C$ is the concentration of the reactant, $M_T = R\sqrt{k/D}$ is known as Thiele's modulus and $k$ and $D$ represent the reaction rate constant and diffusivity of the reacting species. The boundary conditions for this problem is \[C(R) = C_s.\] The analytical solution can be found easily using cylindrical coordinates and is given by \begin{equation} \frac{C(r)}{C_S} = \frac{I_0(r M_T)}{I_0(R M_T)}, \end{equation} where $I_0(r)$ is the modified Bessel function of first kind (this function is available in the library Boost as well as scripting languages such as Python or MATLAB). The conversion from cartesian to cylindrical coordinates is given by \[r = \sqrt{x^2+y^2}.\]
Error indicators
To compare the quality of the refined meshes for the described problem case we look at different error criteria including the max norm $L_\infty$ defined as \begin{equation} L_\infty = \mathrm{max}_i \left|C_i^\mathrm{numerical} - C_i^\mathrm{analytical}\right|, \end{equation} the $L_2$ norm per node defined as \begin{equation} \bar{L_2} = \frac{\sqrt{\sum^N_{i = 1}\left(C_i^\mathrm{numerical} - C_i^\mathrm{analytical}\right)^2}}{N}, \end{equation} where $N$ is the number of nodes (and pertinent equations) in the domain.
We also measure the number of iterations required by the sparse BiCGSTAB solver to reach convergence and the estimated error of solving the system of equations.
Closest node
For a given node $\b{x}_0 = (x_0,y_0)$:
- find the closest node $\b{x}_1 = (x_1,y_1)$
- calculate the half distance between the two nodes \[d = |\b{x}_1 - \b{x}_0|/2\]
- randomly select up to 6 (The case of 6 nodes is the limit since it produces a regular hexagon. In practice this never occurs due to the "monte carlo" node selection procedure.) new nodes on the circle with center $\b{x}_0$ and radius $d$ and simultaneously make sure their is a minimal inter-nodal distance $d$ between the new nodes.
For boundary points we first select 2 points that intersect with the boundary of the domain and only then points lying inside the domain. Due to geometrical constraints boundary points will usually end up with 3 new nodes (in case of straight boundaries we could end up with 4, which would be the previously discussed hexagon limit).
Average radius
Input parameters: $f$ and $l_s$
For a given node $\b{x}_0$:
- find the $l_s$ (an integer from 1 to 7) closest nodes
- calculate the average distance $\bar{d}$ to the $l_s$ closest nodes
- randomly select up to 6 new nodes on the circle with center $\b{x}_0$ and radius $f\cdot\bar{d}$ where $f$ is the radius fraction that lies between 0.2 (leads to clustering) and 0.8. Only allow nodes that are separated by the distance $f \cdot \bar{d}$.
(note that in case $l_s = 1$ and $f = 0.5$ the average radius mechanism becomes equal to the closest node refinement approach described above)
Half-links
Input parameters: $l_s$, $d_m$
For a given node $\b{x}_0$:
- find the $l_s$ (an integer from 1 to 7) closest nodes $\b{x}_i$
- select new nodes in the middle of the segments $\b{x}_i - \b{x}_0$ only allowing points that are separated by the minimal distance $d_m$
(note also that in the 1D case the half-link and closest radius approach become the same)
The minimal distance $d_m$ is chosen as a fraction of the distance to the closest link, e.g. $d_m = f d$, where $f$ is the provided fraction and $d$ is the distance to the closest link.
After experimentation we noticed there are some inconsistencies when trying to refine structured point sets with this approach. The reason for these inconsistencies is that the boundary and internal points have a different number of "natural neighbours". For example in 2D on a square grid, the internal points have 8 neighbours, while boundary points have 5 neighbours. If we choose higher numbers e.g 9 links for an internal node, the 9th node might be any of the 4 nodes one shell further out that only differ at machine precision.
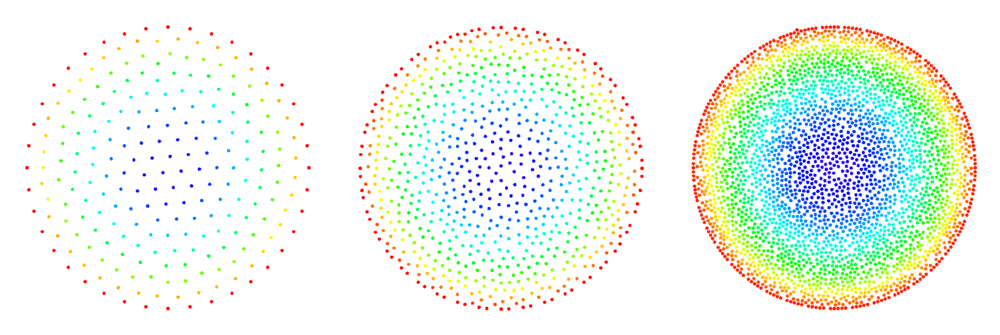
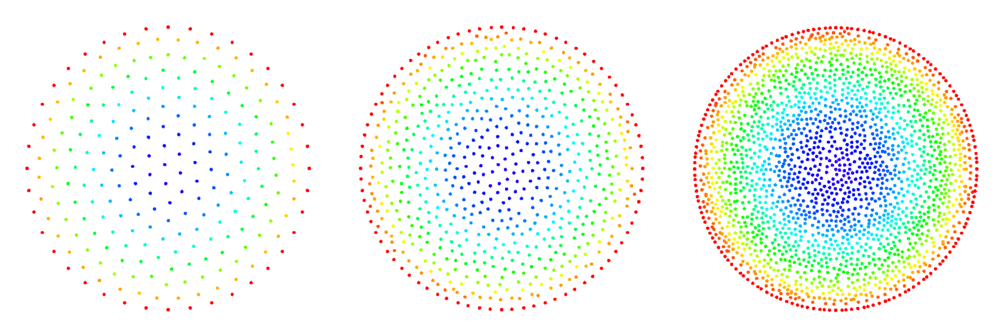
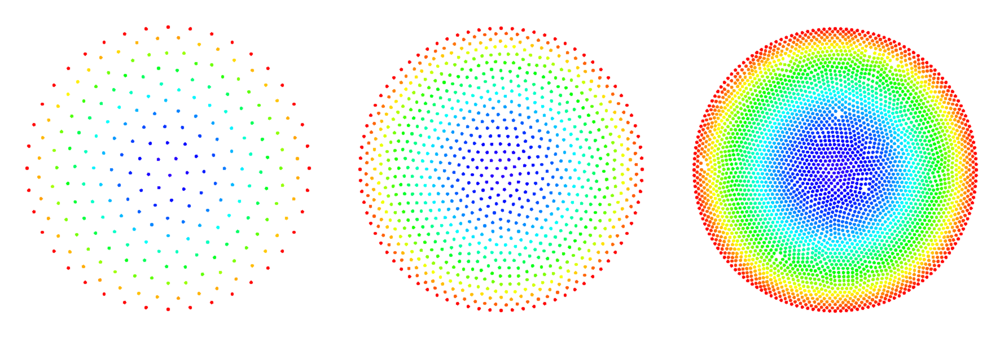
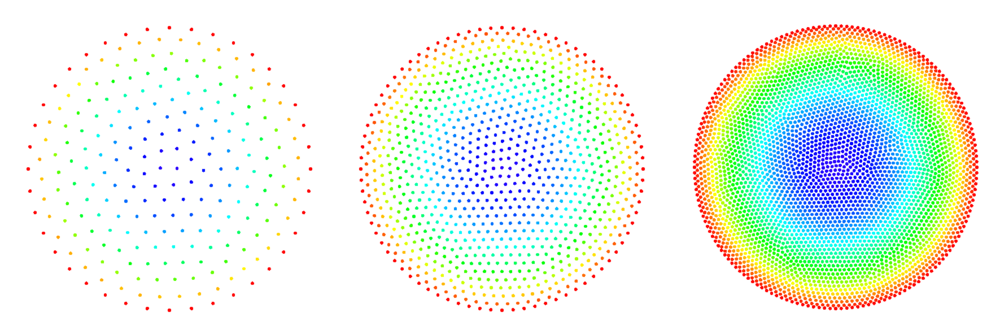
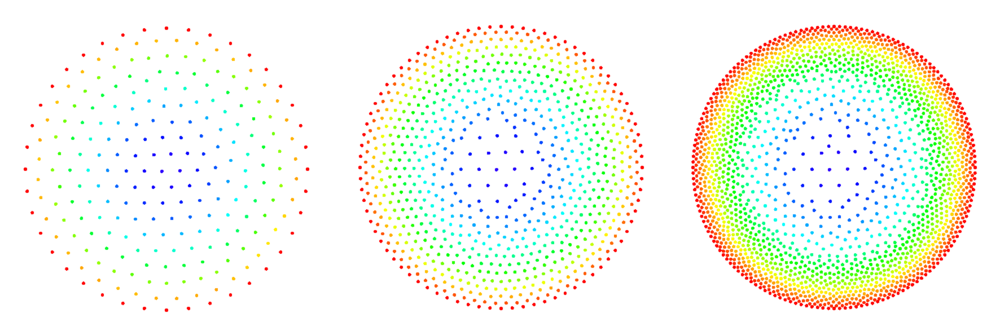
The figures below show some preliminary results of refinement based on half-links. For the circle domain relaxation was applied after the refinement.
After the second refinement of the corner, the solver had difficulty converging to the solution. This was the result of a fixed size shape parameter in the shape functions of the node points. The shape functions have to be tailored to the local characteristic distance in the point set.
Hybrid approach
A hybrid might give better distributions of the refined points. The half-link approach performs well at the boundaries while the distance approach gives less regular internal distributions. In any case it is suggested to perform a few more relaxation steps to equilibrate the mesh.