Difference between revisions of "Analysis of MLSM performance"
(→Solving in 3D) |
(→Solving mixed boundary conditions with MLSM operators) |
||
| Line 115: | Line 115: | ||
<figure id="fig:mikimouse_heat"> | <figure id="fig:mikimouse_heat"> | ||
| − | [[File:mikimouse_heat.png|thumb|upright=2 | + | [[File:mikimouse_heat.png|thumb|upright=2|<caption>Miki mouse domain </caption>]] |
</figure> | </figure> | ||
Revision as of 18:20, 2 February 2017
Contents
Solving Diffusion equation
For starters, we can solve simple Diffusion equation $ \nabla^2 u = \frac{\partial u}{\partial t} $.
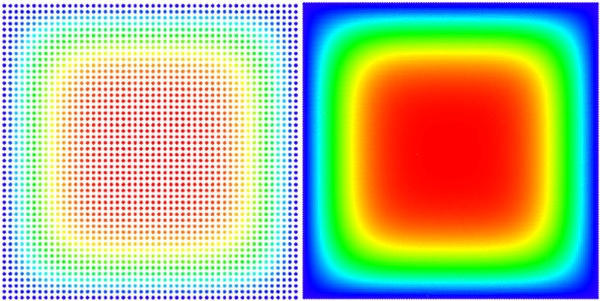
We solved the equation on a square $\Omega = [0, a] \times [0, a]$ with Dirichlet boundary conditions $ \left. u\right|_{\partial \Omega} = 0 $ and initial state $ u(t = 0) = 1$.
An analytical solution for this domain is known, and we use it to evaluate or own solution. \begin{equation} u(\vec{p}, t) = \sum_{\substack{n=1 \\ n \text{ odd}}}^\infty\sum_{\substack{m=1 \\ m \text{ odd}}}^\infty \frac{1}{\pi^2} \frac{16 a^2}{nm} \sin\left(\frac{\pi n}{a}p_x\right) \sin\left(\frac{\pi m}{a}p_y\right) e^{-\frac{\pi^2 (n^2+m^2)}{a^2}t} \end{equation} Because the solution is given in the series form, we only compare to the finite approximation, summing to $N = 100$ instead of infinity. Solution is on Figure 1. See the code for solving diffusion here.
Analysis of MLSM
Convergence with respect to number of nodes
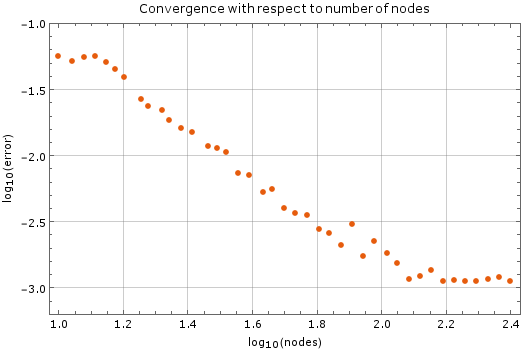
We tested the method with a fixed time step of $ \Delta t = 1\cdot 10^{-5}$ on a unit square ($a = 1$). Results are on Figure 2. Monomial basis of $6$ monomials was used and $12$ closest nodes counted as support for each node. After more than $250$ nodes of discretization in each dimension the method diverges, which is expected. The stability criterion for diffusion equation in two dimensions is $\Delta t \leq \frac{1}{4} \Delta x^2$, where $\Delta x$ is the spatial discretization step in one dimension. In our case, at $250$ nodes per side, the right hand side yields $\frac{1}{4}\cdot\frac{1}{250}\cdot\frac{1}{250} = 4\times 10^{-6}$, so our method is stable within the expected region.
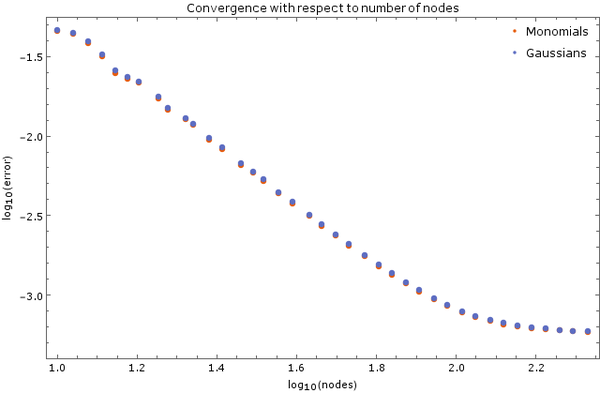
On Figure 3 is another image of convergence, this time using monomial basis $\{1, x, y, x^2, y^2\}$ and Gaussian basis with discretization step $ \Delta t = 5 \cdot 10^{-6}$ and 5 support nodes. Total node count was $N = 2500$. Error was calculated after $0.01$ time units have elapsed.
Convergence with respect to number of time steps
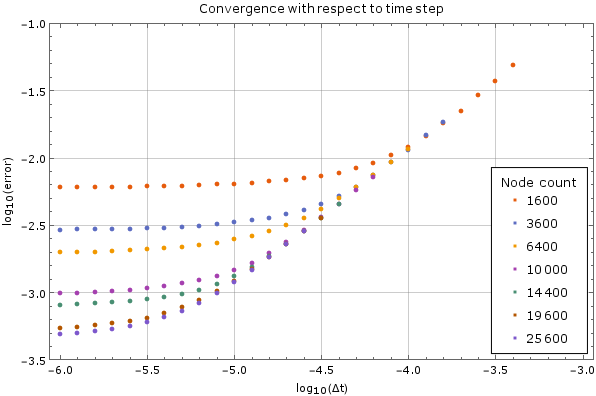
We tested the method on a fixed node count with different time steps on the same domain as above. Results are on Figure 4. For large time steps the method diverges, but once it starts converging the precision increases steadily as the time step decreases, until it hits its lower limit. This behaviour is expected. The error was calculated against the analytical solution above after $0.005$ units of time have passed. A monomial basis up to order $2$ inclusive ($m = 6$) was used and the support size was $n = 12$.
Using Gaussian basis
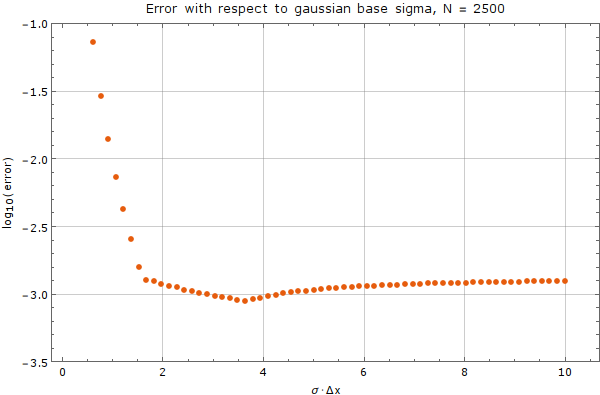
We tested the method on a fixed node count of $N = 2500$ with spatial step discretization of $\Delta x = \frac{1}{50}$. We used the Gaussian basis of $m = 5$ functions with support size $n = 13$. Error was calculated against analytical solution above after $0.01$ time units. A time step of $\Delta t = 10^{-5}$ was used. Results are on Figure 5
As we can see from the graph, there exists an interval where the choice of $\sigma$ does not matter much, but outside of this interval, the method diverges rapidly. Care from the user side must be taken, to choose $\sigma$ appropriately, with respect to plot above.
Solving in 3D
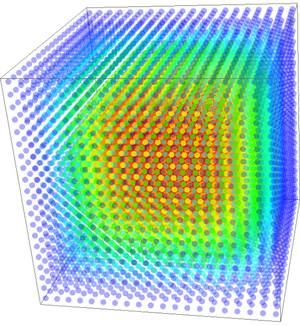
A 3-dimensional case on domain $[0, 1]^3$ was tested on $N = 20^3$ nodes, making the discretization step $\Delta x = 0.05$. Support size of $n=10$ with $m=10$ Gaussian basis functions was used. Their normalization parameter was $\sigma = 60\Delta x$. A time step of $\Delta t = 10^{-5}$ and an explicit Euler method was used to calculate the solution of to $0.01$ time units. Resulting function is on Figure 6.
Solving Dirichlet with EngineMLS
Example code using explicit stepping to reproduce the thermal images from the beginning: example code
Solving Dirichlet with MLSM operators
Example code using explicit stepping and MLSM operators to reproduce the thermal images from the beginning: example code
Solving mixed boundary conditions with MLSM operators
By using MLSM operators and utilizing its `MLSM::neumann` method we can also solve partial diffusion equations. Solution is on Figure 7.
Example code showing the use of Neumann boundary conditions: example code
We also support more -- interesting -- domains :) On Figure 8 we see a domain in a shape of Miki mouse.
Solving Electrostatics
This example is taken from the FreeFem++ manual (page 235).
Assuming there is no current and the charge distribution is time independent, the electric field $\b{E}$ satisfies \begin{equation}\label{eq:electrostatics} \b{\nabla}\cdot\b{E} = \frac{\rho}{\epsilon}, \quad \b{\nabla} \times \b{E} = 0 \end{equation} where $\rho$ is the charge density and $\epsilon$ is the permittivity. If we introduce an electrostatics potential $\phi$ such that \begin{equation} \b{E} = -\b{\nabla}\phi, \end{equation} we can insert it into the first equation in (\ref{eq:electrostatics}) resulting in Poisson's equation \begin{equation}\label{eq:poisson} \b{\nabla}^2 \phi = -\frac{\rho}{\epsilon}. \end{equation} In the absence of unpaired electric charge equation (\ref{eq:poisson}) becomes Laplace's equation \begin{equation} \b{\nabla}^2 \phi = 0 \end{equation}
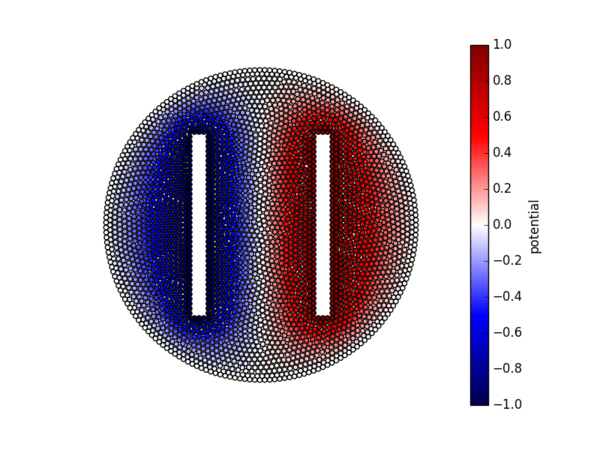
We now solve this equation for a circular enclosure with two rectangular holes. The boundary of the circular enclosure is held at constant potential $0$ V. The two rectangular holes are held at constant potentials $+1$ V and $-1$ V, respectively. A coloured scatter plot is available in figure Figure 9.
1 // domain parameters, size, support and monomial order
2 int domain_size = 3000;
3 size_t n = 15;
4 size_t m = 3;
5
6 double radius = 5.0;
7 double width = 0.3;
8 double height = 3.0;
9
10 // extra boundary labels
11 int LEFT = -10;
12 int RIGHT = -20;
13
14 // build circular domain
15 CircleDomain<Vec2d> domain({0,0},radius);
16 domain.fillUniformInterior(domain_size);
17 domain.fillUniformBoundaryWithStep(domain.characteristicDistance());
18
19 // build left rectangle
20 RectangleDomain<Vec2d> left({-2-width,-height},{-2+width,height});
21 left.fillUniformBoundaryWithStep(domain.characteristicDistance());
22 left.types[left.types == BOUNDARY] = LEFT;
23 domain.subtract(left);
24
25 // build right rectangle
26 RectangleDomain<Vec2d> right({2-width,-height},{2+width,height});
27 right.fillUniformBoundaryWithStep(domain.characteristicDistance());
28 right.types[right.types == BOUNDARY] = RIGHT;
29 domain.subtract(right);
30
31 // relax domain and find supports
32 domain.relax(50, 1e-2, 1.0, 3, 1000);
33 domain.findSupport(n);
34
35 // get interior and boundary ranges
36 Range<int> interior = domain.types == INTERNAL;
37 Range<int> boundary = domain.types == BOUNDARY;
38 Range<int> leftrect = domain.types == LEFT;
39 Range<int> rightrect = domain.types == RIGHT;
40
41 // initialize unknown concentration field and right-hand side
42 VecXd phi(domain.size(),1);
43 VecXd RHS(domain.size(),1);
44
45 // initialize interior values (this is an important step)
46 RHS[interior] = 0.0;
47
48 // set dirichlet boundary conditions
49 RHS[boundary] = 0.0;
50 RHS[leftrect] = -1.0;
51 RHS[rightrect] = 1.0;
52
53 // prepare shape functions and laplacian
54 std::vector<Triplet<double>> l_coeff;
55 for (auto& c : interior) {
56 Range<Vec2d> supp_domain = domain.positions[domain.support[c]];
57 EngineMLS<Vec2d, Monomials, Gaussians> MLS(m,supp_domain,pow(domain.characteristicDistance(),2));
58 VecXd shape = MLS.getShapeAt(supp_domain[0], {{2, 0}}) +
59 MLS.getShapeAt(supp_domain[0], {{0, 2}});
60 for (size_t i = 0; i < supp_domain.size(); ++i) {
61 l_coeff.emplace_back(c, domain.support[c][i], shape[i]);
62 }
63 }
64
65 // prepare dirichlet boundaries
66 std::vector<Triplet<double>> b_coeff;
67 for (auto& c : domain.types < 0) {
68 b_coeff.emplace_back(c, c, 1.0);
69 }
70
71 // prepare matrices
72 SparseMatrix<double> laplace_m(domain.size(), domain.size());
73 laplace_m.setFromTriplets(l_coeff.begin(), l_coeff.end());
74 SparseMatrix<double> boundary_m(domain.size(), domain.size());
75 boundary_m.setFromTriplets(b_coeff.begin(), b_coeff.end());
76
77 // draw
78 std::thread th([&] { draw2D(domain.positions, phi); });
79
80 // initialize solver
81 Eigen::BiCGSTAB<SparseMatrix<double>> solver;
82
83 // join matrices together
84 SparseMatrix<double> tmp = boundary_m + laplace_m;
85 solver.compute(tmp);
86
87 // solve system of equations
88 phi = solver.solve(RHS);
89 std::cout << "#iterations: " << solver.iterations() << std::endl;
90 std::cout << "estimated error: " << solver.error() << std::endl;
91
92 // end drawing
93 th.join();
Convection-diffusion
The following example problem is adapted from Ferziger & Perič (2002) (pages 82-86).
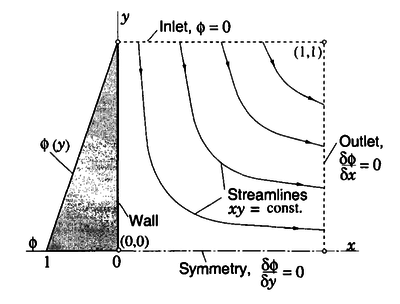
We now consider the problem of a scalar quantity transported in a known velocity field $\b{u}$. The velocity field is given by \[\b{u} = (u_x,u_y) = (x, -y),\] which represents the flow near a stagnation point $(x,y) = (0,0)$. The streamlines $xy=\mathrm{const.}$ and change direction with respect to the Cartesian grid. The convection-diffusion equation (also known as the scalar transport equation) to be solved is \[\alpha \b{\nabla}^2 \phi - \b{u}\cdot \b{\nabla}\phi = 0\] with the following boundary conditions:
- $\phi = 0$ along the north (inlet) boundary;
- linear variation $\phi = (1 - y)$ along the west boundary ($x = 0$);
- symmetry condition on the south boundary;
- zero gradient at the east (outlet) boundary.
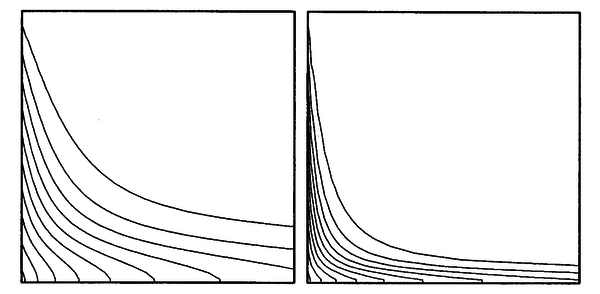
The parameter $\alpha$ is the diffusivity for the scalar quantity $\phi$. The problem and boundary conditions are schematically represented in the Figure 10. The solutions obtained by Ferziger and Perić are shown in Figure 11.
The meshless solution is obtained on regular point arrangements of sizes 100 × 100 and 300 × 300, for the 2 cases of diffusivity $\alpha = 0.01$ and $\alpha = 0.001$, respectively. For comparison we have plotted the contour lines at the same intervals as the original authors of this example (see Figure 12). The basis functions are the 5 monomials $1$, $x$, $y$, $x^2$, $y^2$.